Notes on the Stirling Cycle
In this post I share some notes about the Stirling cycle with derivations of quantities for each process along with code to plot P-V and S-T diagrams for a numerical example.
Contents
- Preliminaries
- Equations overview
- Stirling cycle
- Process $1 \rightarrow 2$
- Process $2 \rightarrow 3$
- Process $3 \rightarrow 4$
- Process $4 \rightarrow 1$
- Efficiency
- Summary
Preliminaries
from IPython.display import display
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pickle
import io
# A function to copy a matplotlib figure (adapted from https://stackoverflow.com/questions/45810557/copy-an-axes-content-and-show-it-in-a-new-figure)
def copy_fig(fig):
buf = io.BytesIO()
pickle.dump(fig, buf)
buf.seek(0)
return pickle.load(buf)
Equations overview
From experimental observations of ideal gases we have the following:
- P-V-T relation: $PV = mRT$ (equation of state)
- If $V$ is constant during a heat transfer $Q$, the gas behaves like a solid: $Q_{12} = mc_V(T_2 - T_1)$
- If the gas is subject to a constant temperature expansion / compression, we observe that $Q_{12} = W_{12}$
From which the following constitutive relations may be derived for energy and entropy
- Constant volume heat transfer:
- Entropy change for a reversible process:
\(\Delta S = mc_V\ln\frac{T_2}{T_1} + mR\ln{\frac{V_2}{V_1}}\)
- P-V relation for isentropic process:
Efficiency (where $Q_C$ and $Q_H$ stand for the heat transferred out of and into the system, respectively). \(\eta = \frac{W_\text{net}}{Q_H}\)
Stirling cycle
- Consists of four reversible processes
- For each process we want to determine
- Pressure volume (P-V) relation
- Change in energy: $\Delta E$
- Heat transfer: $Q$
- Work: $W$
- Change in entropy: $\Delta S$
- The constituents of $\Delta S$:
- Entropy transferred: $S_\text{trans}$
- Entropy generated: $S_\text{gen}$
- Entropy temperatue (S-T) relation
- Since all the processes are reversible no entropy is generated so $S_\text{gen} = 0$
Process $1 \rightarrow 2$
- Isothermal: $T_2 = T_1$
- Expansion at $T_H = T_1$
Note that this is identical to process $1\rightarrow 2$ of the Carnot cycle
P-V relation
- From equation of state
1st law
- Since constant temperature expansion, from observation 3: \(\Delta E = 0\) \(Q_{12} = W_{12} = \int_1^2 P dV = \int_1^2 \frac{mRT}{V} dV = mRT_1\ln{\frac{V_2}{V_1}}\)
2nd law
- Because of reversible and isothermal nature of the process, ${S_\text{gen}}$
- Since $S_\text{gen} = 0$
Summary
- $P = \frac{mRT_1}{V}$
- $\Delta E = 0$
- $Q_{12} = mRT_1\ln{\frac{V_2}{V_1}}$
- $W_{12} = mRT_1\ln{\frac{V_2}{V_1}}$
- $\Delta S = mR\ln{\frac{V_2}{V_1}}$
- $S_\text{trans} = mR\ln{\frac{V_2}{V_1}}$
- $S_\text{gen} = 0$
- On the $S-T$ diagram, since $T$ is constant, there is a straight horizontal line $T = T_1$ between $S_1$ and $S_2 = S_1 + mR\ln{\frac{V_2}{V_1}}$
Example
Some quanities for a cycle
cv = 718
R = 287
TH = 600
TC = 500
V1 = 0.01
V2 = 0.02
P1 = 1e6
We can derive other quantities from the above
T1 = TH
T2 = T1 # isothermal
# state equation to find P2 and m
m = P1 * V1 / (R * T1)
P2 = m*R*T2/V2
print(f'm = {m}')
print(f'P2 = {P2}')
m = 0.05807200929152149
P2 = 500000.0
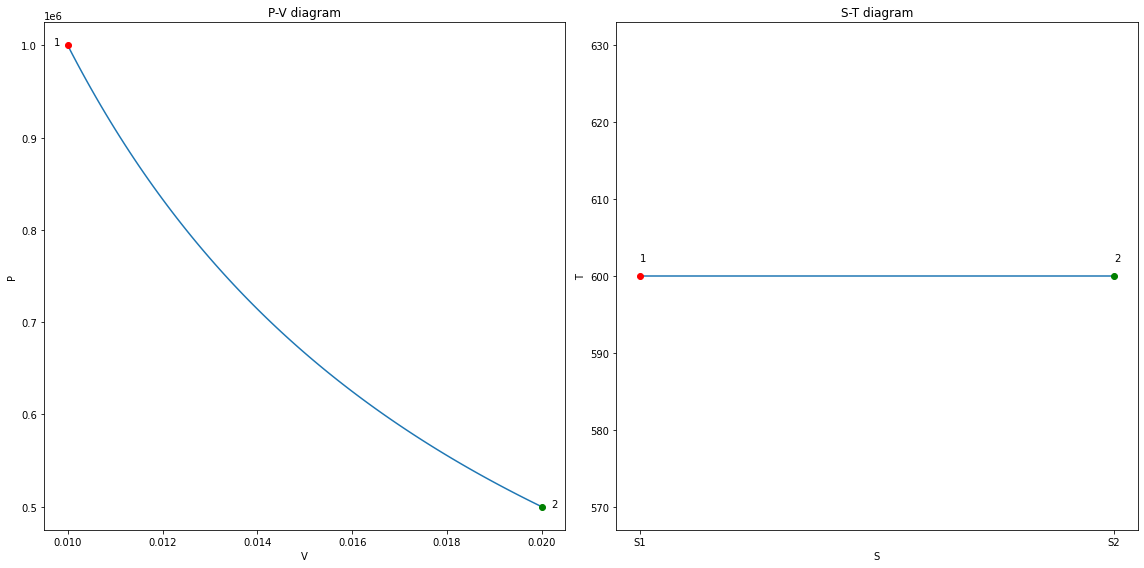
The P-V and S-T plots
V = np.linspace(V1, V2, 101)
P = m*R*T1 / V
assert np.isclose(P[-1], P2)
fig12, (ax1, ax2) = plt.subplots(1, 2, figsize=(16, 8))
ax1.plot(V, P)
line_clr = ax1.lines[0].get_color()
ax1.plot(V1, P1, marker='o', color='red')
ax1.plot(V2, P2, marker='o', color='green');
ax1.set_xlabel('V')
ax1.set_ylabel('P')
ax1.set_title('P-V diagram')
ax1.text(V1-3e-4, P1, '1')
ax1.text(V2 + 2e-4, P2, '2')
S1 = 1 # Some arbitrary value
S2 = S1 + m*R*np.log(V2/V1)
ax2.plot(np.linspace(S1, S2, 101), np.ones(101)*T1)
ax2.plot(S1, T1, marker='o', color='red')
ax2.plot(S2, T2, marker='o', color='green');
ax2.text(S1, T1 + 2, '1')
ax2.text(S2, T2 + 2, '2')
ax2.set_xticks([S1, S2]);
ax2.set_xticklabels(['S1', 'S2']);
ax2.set_xlabel('S')
ax2.set_ylabel('T');
ax2.set_title('S-T diagram');
fig12.tight_layout();
state_df1 = pd.DataFrame(
{'state': [1, 2],
'P': [P1, P2],
'V': [V1, V2],
'T': [T1, T2]}
)
Q12 = m*R*T1*np.log(V2/V1)
W12 = Q12
dE = Q12 - W12
dS12 = S2 - S1
S_trans_12 = dS12
S_gen_12 = 0
process_df1 = pd.DataFrame(
{'process': ['1 → 2'],
'ΔE': [dE],
'Q': [Q12],
'W': [W12],
'ΔS': [dS12],
'S_trans': [S_trans_12],
'S_gen': [S_gen_12]
}
)
display(state_df1.round(5))
display(process_df1.round(5))
| state | P | V | T | |
|---|---|---|---|---|
| 0 | 1 | 1000000.0 | 0.01 | 600 |
| 1 | 2 | 500000.0 | 0.02 | 600 |
| process | ΔE | Q | W | ΔS | S_trans | S_gen | |
|---|---|---|---|---|---|---|---|
| 0 | 1 → 2 | 0.0 | 6931.47181 | 6931.47181 | 11.55245 | 11.55245 | 0 |
Process $2 \rightarrow 3$
- Ischoric (constant volume)
- Cooling to $T_C = T_3 < T_2$
P-V relation
- From equation of state, with $V_3 = V_2$
1st Law
- Isochoric so $W_{23} = 0$
2nd law
- Since constant volume
Summary
- On the $P-V$ diagram, since $V$ is constant, there is a straight vertical with $P_3 = mRT_3/V_3$ and $P_2$ at each end
- $\Delta E = mc_V(T_3 - T_2)$
- $Q_{23} = mc_V(T_3 - T_2)$
- $W_{23} = 0$
- $\Delta S = mc_V\ln\frac{T_3}{T_2}$
- $S_\text{trans} = mc_V\ln\frac{T_3}{T_2}$
- $S_\text{gen} = 0$
- $S = S_2 + mc_V\ln\frac{T}{T_2}$
Example (continued)
T3 = TC
gamma = 1 + R/cv
print(f'gamma = {gamma}')
V3 = V2
print(f'V3 = {V3}')
P3 = m * R * T3 / V3
print(f'P3 = {P3}')
gamma = 1.3997214484679665
V3 = 0.02
P3 = 416666.6666666667
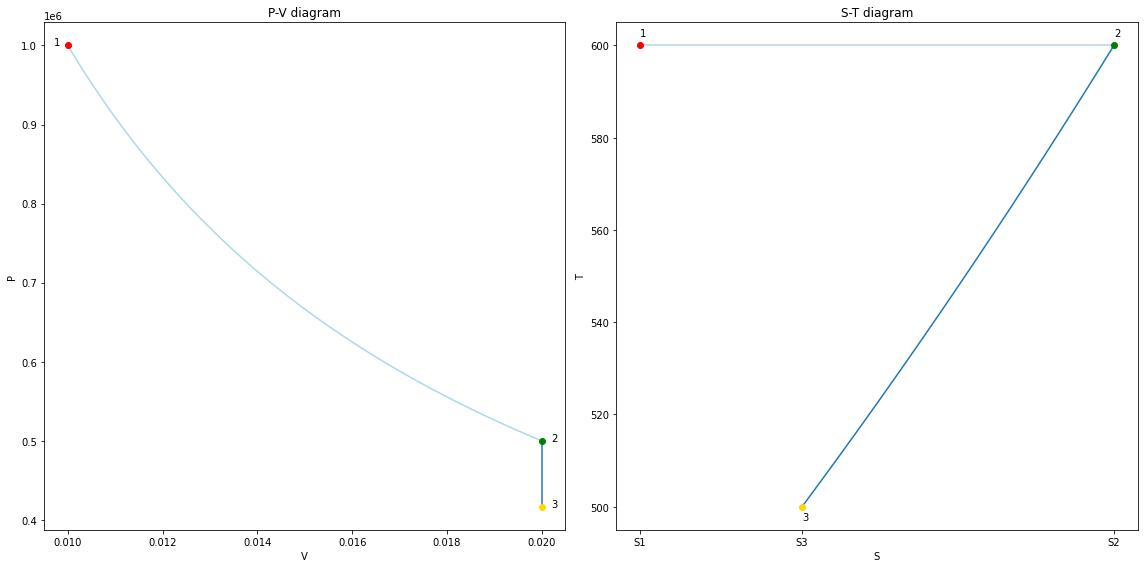
V = np.ones(101) * V2
P = np.linspace(P2, P3, 101)
assert np.isclose(P[-1], P3)
fig23 = copy_fig(fig12)
ax1, ax2 = fig23.get_axes()
for line in ax1.lines + ax2.lines:
if line.get_color() == line_clr:
line.set_color('lightblue')
ax1.plot(V, P, color=line_clr, zorder=-1)
ax1.plot(V3, P3, marker='o', color='gold');
ax1.text(V3 + 2e-4, P3, '3')
T = np.linspace(T2, T3, 101)
S = S2 + cv * m * np.log(T/T2)
S3 = S2 + cv * m * np.log(T3/T2)
assert np.isclose(S[-1], S3)
ax2.plot(S, T, color=line_clr, zorder=-1)
ax2.plot(S3, T3, marker='o', color='gold');
ax2.set_xticks([S1, S2, S3]);
ax2.set_xticklabels(['S1', 'S2', 'S3']);
ax2.text(S3, T3 - 3, '3')
fig23.tight_layout();
fig23
state_df2 = pd.concat(
[state_df1,
pd.DataFrame(
{'state': [3],
'P': [P3],
'V': [V3],
'T': [T3]}
)], axis=0
).reset_index(drop=True)
Q23 = m*cv*(T3 - T2)
W23 = 0
dE = Q23 - W23
dS23 = S3 - S2
S_trans_23 = dS23
S_gen_23 = 0
process_df2 = pd.concat(
[process_df1,
pd.DataFrame(
{'process': ['2 → 3'],
'ΔE': [dE],
'Q': [Q23],
'W': [W23],
'ΔS': [dS23],
'S_trans': [S_trans_23],
'S_gen': [S_gen_23]
})
], axis=0
)
display(state_df2.round(5))
display(process_df2.round(5))
| state | P | V | T | |
|---|---|---|---|---|
| 0 | 1 | 1000000.00000 | 0.01 | 600 |
| 1 | 2 | 500000.00000 | 0.02 | 600 |
| 2 | 3 | 416666.66667 | 0.02 | 500 |
| process | ΔE | Q | W | ΔS | S_trans | S_gen | |
|---|---|---|---|---|---|---|---|
| 0 | 1 → 2 | 0.00000 | 6931.47181 | 6931.47181 | 11.55245 | 11.55245 | 0 |
| 0 | 2 → 3 | -4169.57027 | -4169.57027 | 0.00000 | -7.60203 | -7.60203 | 0 |
Process $3 \rightarrow 4$
- Isothermal
- Compression at $T_C = T_3$
The derivations are analogous to $1 \rightarrow 2$ so we can just need to replace the process ids in the results obtained earlier.
Note also that this is too is identical to process $3 \rightarrow 4$ of the Carnot cycle
Summary
- $P = \frac{mRT_3}{V}$
- $\Delta E = 0$
- $Q_{34} = mRT_3\ln{\frac{V_4}{V_3}}$
- $W_{34} = mRT_3\ln{\frac{V_4}{V_3}}$
- $\Delta S = mR\ln{\frac{V_4}{V_3}}$
- $S_\text{trans} = mR\ln{\frac{V_4}{V_3}}$
- $S_\text{gen} = 0$
- On the $S-T$ diagram, since $T$ is constant, there is a straight horizontal line $T = T_3$ between $S_3$ and $S_4=S_3 + mR\ln{\frac{V_4}{V_3}}$
Example (continued)
Note that to for $V_4$ we use the fact that the next process is isochoric
T4 = T3
V4 = V1
P4 = m*R*T4/V4
print(f'V4 = {V4}')
print(f'P4 = {P4}')
V4 = 0.01
P4 = 833333.3333333334
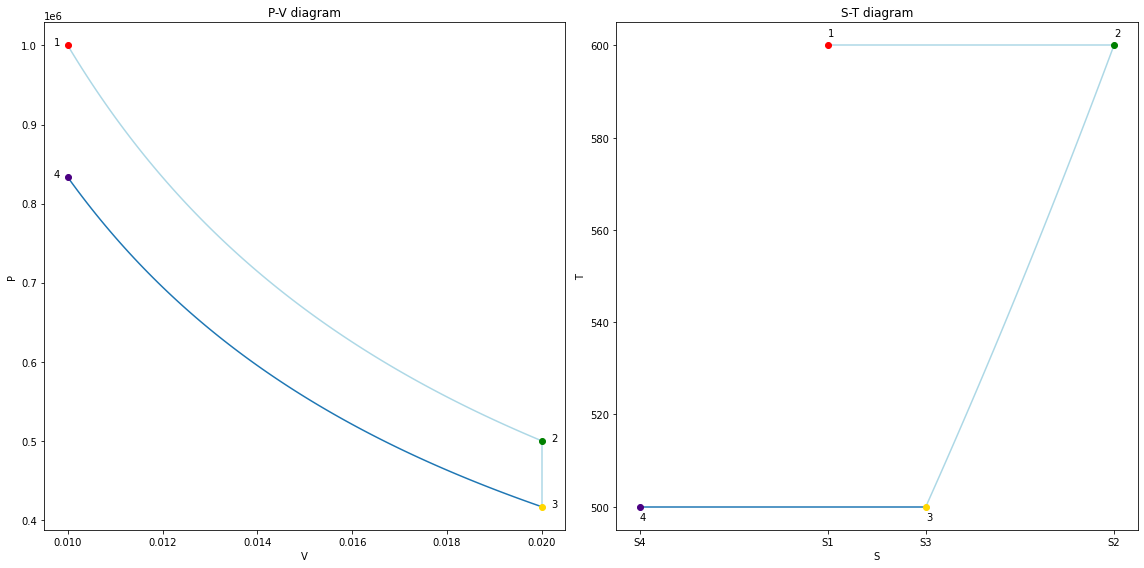
V = np.linspace(V3, V4, 101)
P = m*R*T3/V
assert np.isclose(P[-1], P4)
fig34 = copy_fig(fig23)
ax1, ax2 = fig34.get_axes()
for line in ax1.lines + ax2.lines:
if line.get_color() == line_clr:
line.set_color('lightblue')
ax1.plot(V, P, color=line_clr, zorder=-1)
ax1.plot(V4, P4, marker='o', color='indigo');
ax1.text(V4 - 3e-4, P4, '4')
S4 = S3 + m*R*np.log(V4/V3)
ax2.plot(np.linspace(S3, S4, 101), np.ones(101)*T3, color=line_clr, zorder=-1)
ax2.plot(S4, T4, marker='o', color='indigo');
ax2.text(S4, T4 - 3, '4')
ax2.set_xticks([S1, S2, S3, S4]);
ax2.set_xticklabels(['S1', 'S2', 'S3', 'S4']);
fig34.tight_layout();
fig34
state_df3 = pd.concat(
[state_df2,
pd.DataFrame(
{'state': [4],
'P': [P4],
'V': [V4],
'T': [T4]}
)], axis=0
).reset_index(drop=True)
Q34 = m*R*T3*np.log(V4/V3)
W34 = Q34
dE = Q34 - W34
dS34 = S4 - S3
S_trans_34 = dS34
S_gen_34 = 0
process_df3 = pd.concat(
[process_df2,
pd.DataFrame(
{'process': ['3 → 4'],
'ΔE': [dE],
'Q': [Q34],
'W': [W34],
'ΔS': [dS34],
'S_trans': [S_trans_34],
'S_gen': [S_gen_34]
})
], axis=0
)
display(state_df3.round(5))
display(process_df3.round(5))
| state | P | V | T | |
|---|---|---|---|---|
| 0 | 1 | 1000000.00000 | 0.01 | 600 |
| 1 | 2 | 500000.00000 | 0.02 | 600 |
| 2 | 3 | 416666.66667 | 0.02 | 500 |
| 3 | 4 | 833333.33333 | 0.01 | 500 |
| process | ΔE | Q | W | ΔS | S_trans | S_gen | |
|---|---|---|---|---|---|---|---|
| 0 | 1 → 2 | 0.00000 | 6931.47181 | 6931.47181 | 11.55245 | 11.55245 | 0 |
| 0 | 2 → 3 | -4169.57027 | -4169.57027 | 0.00000 | -7.60203 | -7.60203 | 0 |
| 0 | 3 → 4 | 0.00000 | -5776.22650 | -5776.22650 | -11.55245 | -11.55245 | 0 |
Process $4 \rightarrow 1$
- Isochoric
- Heating to $T_H = T_1$
The derivations are analogous to $2 \rightarrow 3$ so we can just need to replace the process ids in the results obtained earlier
Summary
- On the $P-V$ diagram, since $V$ is constant, there is a straight vertical with $P_1 = mRT_1/V_1$ and $P_4$ at each end
- $\Delta E = mc_V(T_1 - T_4)$
- $Q_{41} = mc_V(T_1 - T_4)$
- $W_{41} = 0$
- $\Delta S = mc_V\ln\frac{T_4}{T_1}$
- $S_\text{trans} = mc_V\ln\frac{T_4}{T_1}$
- $S_\text{gen} = 0$
- $S = S_4 + mc_V\ln\frac{T}{T_4}$
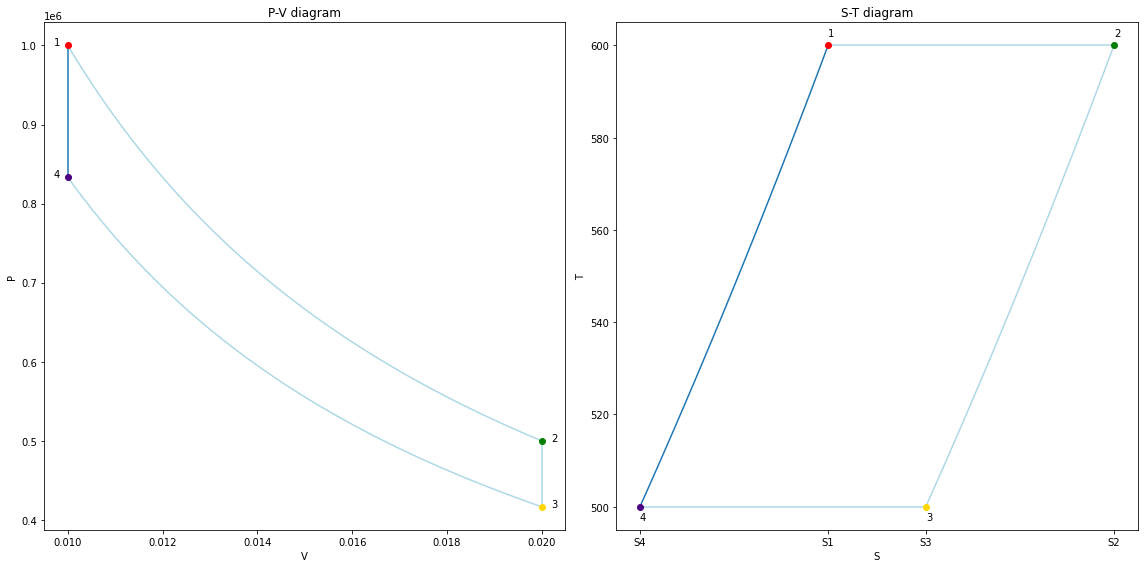
V = np.ones(101) * V4
P = np.linspace(P4, P1, 101)
assert np.isclose(P[-1], P1)
fig41 = copy_fig(fig34)
ax1, ax2 = fig41.get_axes()
for line in ax1.lines + ax2.lines:
if line.get_color() == line_clr:
line.set_color('lightblue')
ax1.plot(V, P, color=line_clr, zorder=-1)
T = np.linspace(T4, T1, 101)
S = S4 + cv * m * np.log(T/T4)
assert np.isclose(S[-1], S1)
ax2.plot(S, T, color=line_clr, zorder=-1)
fig41
Q41 = m*cv*(T1 - T4)
W41 = 0
dE = Q41 - W41
dS41 = S1 - S4
S_trans_41 = dS41
S_gen_41 = 0
process_df4 = pd.concat(
[process_df3,
pd.DataFrame(
{'process': ['4 → 1'],
'ΔE': [dE],
'Q': [Q41],
'W': [W41],
'ΔS': [dS41],
'S_trans': [S_trans_41],
'S_gen': [S_gen_41]
})
], axis=0
)
display(state_df3.round(5))
display(process_df4.round(5))
| state | P | V | T | |
|---|---|---|---|---|
| 0 | 1 | 1000000.00000 | 0.01 | 600 |
| 1 | 2 | 500000.00000 | 0.02 | 600 |
| 2 | 3 | 416666.66667 | 0.02 | 500 |
| 3 | 4 | 833333.33333 | 0.01 | 500 |
| process | ΔE | Q | W | ΔS | S_trans | S_gen | |
|---|---|---|---|---|---|---|---|
| 0 | 1 → 2 | 0.00000 | 6931.47181 | 6931.47181 | 11.55245 | 11.55245 | 0 |
| 0 | 2 → 3 | -4169.57027 | -4169.57027 | 0.00000 | -7.60203 | -7.60203 | 0 |
| 0 | 3 → 4 | 0.00000 | -5776.22650 | -5776.22650 | -11.55245 | -11.55245 | 0 |
| 0 | 4 → 1 | 4169.57027 | 4169.57027 | 0.00000 | 7.60203 | 7.60203 | 0 |
Efficiency
\[W_\text{net} = W_{12} + W_{23} + W_{34} + W_{41} \\ = mRT_1\ln{\frac{V_2}{V_1}} + 0 + mRT_3\ln{\frac{V_4}{V_3}} + 0\]Since
\(V_3 = V_2\) \(V_4 = V_1\)
we have
\[\frac{V_3}{V_4} = \frac{V_2}{V_1}\]Hence
\[W_\text{net} = mR(T_1 - T_3)\ln{\frac{V_2}{V_1}} = mR(T_H - T_C)\ln{\frac{V_2}{V_1}}\]Note that we also need to include an additional term heat transfer term $Q_{41}$ which was absent in the Carnot cycle since process $4 \rightarrow 1$ of the Carnot cycle was adiabatic.
\[Q_H = Q_{41} + Q_{12} = mc_V(T_1 - T_4) + mRT_1\ln{\frac{V_2}{V_1}} = mc_V(T_H - T_C) + mRT_H\ln{\frac{V_2}{V_1}}\]which means
\[\eta = \frac{W_\text{net}}{Q_H} = \frac{mR(T_H - T_C)\ln{\frac{V_2}{V_1}}}{mc_V(T_H - T_C) + mRT_H\ln{\frac{V_2}{V_1}}} \\= \frac{1}{\frac{c_V}{R}\frac{1}{\ln{\frac{V_2}{V_1}}} + \frac{T_H}{T_H - T_C} }\]Note that since $V_2 > V_1 \implies \frac{c_V}{R}\ln{\frac{V_2}{V_1}} > 0$
\[\eta_\text{Stirling} = \frac{1}{\frac{c_V}{R}\frac{1}{\ln{\frac{V_2}{V_1}}} + \frac{1}{\eta_\text{Carnot}} } = \frac{\eta_\text{Carnot}}{\eta_\text{Carnot}\frac{c_V}{R}\frac{1}{\ln{\frac{V_2}{V_1}}} + 1 } < \eta_\text{Carnot}\]This reduction in efficiency is due to the fact that heat transfer occurs with a change in temperature in contrast to the purely isothermal heat transfers found in the Carnot cycle.
Summary
fig_final = copy_fig(fig41)
ax1, ax2 = fig_final.get_axes()
for line in ax1.lines + ax2.lines:
if line.get_color() == 'lightblue':
line.set_color(line_clr)
display(fig_final)
display(state_df3.round(5))
display(process_df4.round(5))
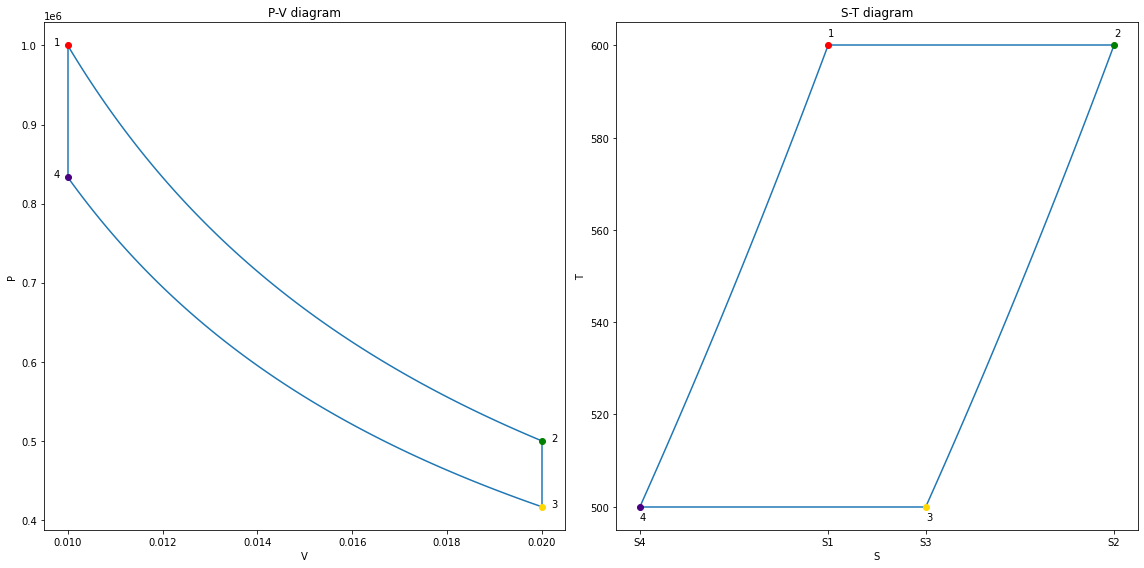
| state | P | V | T | |
|---|---|---|---|---|
| 0 | 1 | 1000000.00000 | 0.01 | 600 |
| 1 | 2 | 500000.00000 | 0.02 | 600 |
| 2 | 3 | 416666.66667 | 0.02 | 500 |
| 3 | 4 | 833333.33333 | 0.01 | 500 |
| process | ΔE | Q | W | ΔS | S_trans | S_gen | |
|---|---|---|---|---|---|---|---|
| 0 | 1 → 2 | 0.00000 | 6931.47181 | 6931.47181 | 11.55245 | 11.55245 | 0 |
| 0 | 2 → 3 | -4169.57027 | -4169.57027 | 0.00000 | -7.60203 | -7.60203 | 0 |
| 0 | 3 → 4 | 0.00000 | -5776.22650 | -5776.22650 | -11.55245 | -11.55245 | 0 |
| 0 | 4 → 1 | 4169.57027 | 4169.57027 | 0.00000 | 7.60203 | 7.60203 | 0 |